Bits, Bytes & Biomolecules:
Bridging Science and Technology to Build the Future
Combining expertise in Bioinformatics and Bioprocesses with AI-driven strategies.
About Me
Currently, as a PhD student in Bioinformatics, I'm deepening my expertise in Machine Learning, Genomics, Transcriptomics, Structural Biology and Chemoinformatics. My research applies deep learning algorithms, focusing on generative models to extract biological insights from complex datasets, with the goal of developing an antimicrobial language for generating synthetic compounds inspired by the immune system of diverse organisms. My ultimate goal is to provide actionable insights for the development of new therapeutics against antimicrobial resistance, using scalable in silico methods.
Simultaneously, I'm also a MBA student in Software Engineering & Data Science, where I'm expanding my expertise in key areas such as data analysis for strategic insights, advanced machine learning, data engineering and governance.
I hold a Master’s degree in Genetics and Molecular Biology, during which I developed the AMPidentifier, an automated machine learning-based tool designed to identify antimicrobial sequences in genomic and protemic data. This tool has shown promising potential in uncovering bioactive compounds in genomes, fostering progress in drug discovery and bioproduct development. Additionally, I hold a Bachelor’s degree in Biomedical Sciences, with a focus on Bioinformatics and Clinical Pathology. I gained practical experience in hospitals and molecular biology research laboratories and further developed my skills in computational biology working as a bioinformatics intern in research centers.
In the private sector, I worked as Research and Development Analyst, focusing on the end-to-end development of IoT and AI products. My responsibilities included building roadmaps, defining KPIs, developing MVPs and leading user feedback collection to guide product upgrades. By leveraging data-driven decision-making and applying machine learning to health data, I delivered efficient user-centric solutions. Using Agile, Lean and Scrum approaches, I worked on optimizing team performance and ensuring a well-designed project delivery. Additionally, I conducted market research to identify trends, collaborated with stakeholders to align goals and ensured continuous improvement through agile retrospectives.
Combining skills in research, product management and agile methods, I feel ready to deliver innovative solutions to real-world challenges in general products and especially in life sciences and healthcare, enhancing technology and user experience.
Skills
Bioinformatics
- Genomics & Transcriptomics
- Structural Genomics
- Genome Mining and Annotation
- Molecular & Structural Biology
- Protein & Membrane Modeling
- Chemoinformatics & Protein Design
- Molecular Dynamics Simulations
- Pipeline Development
AI, Data & Scripting
- Machine Learning (Supervised, Unsupervised, Generative Networks)
- Data Analysis & Visualization
- Data Engineering & Governance
- Python, Bash & SQL
- Linux/Unix Systems
- HPC Workload Management
Product/Project Management
- Product Lifecycle Management
- Agile Methodologies (Scrum, Kanban)
- User-Centered Design (UCD)
- Roadmap Development
- Key Performance Indicators (KPIs)
- Market & User Research
- Lean Product Development / MVP
- Design Thinking
Product Lifecycle Management
- Figma, Miro, Jira, Trello & Azure
- Git & Version Control
- Google Workspace & Notion
- Tableau & PowerBI
Collaboration & Communication
- Cross-Functional Team Leadership
- Stakeholder Management
- Technical & Business Communication
- Complex Problem-Solving
- Data Storytelling
Key Soft Skills
- Active Listening
- Critical Thinking
- Creativity & Innovation
- Teamwork
Experience
Scientific Researcher - PhD Student (Aug 2024 - Now)
Institute of Biosciences (ICB) - Federral University of Minas Gerais (UFMG)
As a PhD student in Bioinformatics at Federral University of Minas Gerais (UFMG), I focus on deep learning, genomics, and structural biology, specifically in the characterization and design of antimicrobial peptides. My research involves developing computational models for membrane interactions and conducting molecular dynamics simulations to enhance peptide stability and functionality, with a focus on developing future candidates for antimicrobials against antimicrobial resistance.
Scientific Researcher - Master's Student (Jun 2022 - Jul 2024)
Department of Genetics (dGEN) - Federral University of Pernambuco (UFPE)
During my Master's in Genetics and Molecular Biology at the Federal University of Pernambuco (UFPE), I developed a tool called AMP-Identifier, based on machine learning, to identify antimicrobial peptides (AMPs) in genomic sequences. The project involved applying computational techniques to analyze large datasets, enabling the discovery of new molecules and molecular signatures, thus contributing to advancements in bioinformatics and potential therapeutic applications. Additionally, I conducted omics data analyses, enhancing the understanding of peptide functions and their applications in medicine.
Research & Development Analyst (Oct 2020 - Aug 2023)
PickCells
I worked on the development of applications for IoT solutions that use computer vision for pattern recognition in clinically relevant images. My responsibilities included prototyping, developing, and validating systems.
Undergraduate Researcher Fellow (Jan 2020 - May 2022)
Laboratory of Genetics and Vegetal Biotchnology (LGBV) - Federral University of Pernambuco (UFPE)
Conducted research in Bioinformatics aimed at the characterization and optimization of bio-inspired antimicrobial peptides. Additionally, I worked on the investigation of eIF4E sequences in Vigna species, focusing on their potential roles in plant defense mechanisms against pathogens.
Clinical Pathology Intern (Oct 2021 - Mar 2022)
Brazilian Hospital Services Company (EBSERH) - Clinical Hospital of UFPE (HC/PE)
Engaged in activities related to Clinical Pathology within the Clinical Analysis Laboratory at HC/UFPE, specifically in the areas of Sample Screening, Hematology, Biochemistry, Urinalysis, Microbiology, Hormone Testing and Serology. This role involved conducting various diagnostic tests, analyzing results and ensuring quality control within the laboratory, contributing to accurate and timely patient care.
Research, Technological Development and Innovation Intern (Apr 2020 - Aug 2020)
PickCells
Contributed to the development of solutions for the molecular diagnosis of SARS-CoV-2.
Undergraduate Researcher Fellow (Nov 2016 - Dec 2019)
Oswaldo Cruz Foundation (FIOCRUZ)
Engaged in activities in the areas of Structural Biology and Theoretical/Computational Chemistry, focusing on Molecular Modeling and Protein Engineering for diagnostic and vaccine purposes in the Department of Virology and Experimental Therapy.
Undergraduate Researcher Fellow (May 2015 - Dec 2016)
Keizo Asami Institute (iLIKA)
Developed scientific activities in the Molecular Biology sector (in partnership with the Forensic Genetics Institute of the Scientific Police of Pernambuco) in the areas of Human Genetics and Bioinformatics, with an emphasis on Forensic Genetics, Ancestry Markers and Phenotype Prediction.
Education
PhD in Bioinformatics (Ongoing)
Federal University of Minas Gerais (UFMG), Belo Horizonte, Minas Gerais, Brazil
Focusing on machine learning for the identification and design of antimicrobial peptides and the analysis of immune-related biological compounds encoded in multi-omic data. Currently developing scalable in silico methods for the generation of antimicrobial compounds using Transformer-based models.
MBA in Software Engineering (Ongoing)
University of São Paulo (USP), São Paulo, Brazil
Currently studying software engineering principles and practices. The curriculum encompasses full-stack development using Python and JavaScript, microservices architecture, DevOps methodologies, cloud computing with Docker and Kubernetes, information security, and data engineering. The program integrates modern development practices, including agile methodologies, infrastructure as code, and machine learning, equipping professionals to lead complex technological projects and drive organizational innovation.
Postgraduate in Data Science and Analytics (Ongoing)
Pontifical Catholic University of Rio de Janeiro (PUC-Rio), Rio de Janeiro, Brazil
Currently deepening expertise in data-driven decision-making through advanced training in data analysis, statistics and machine learning. This MBA bridges theory and practice, empowering me to extract meaningful insights from complex datasets, build predictive models and apply analytical thinking across real-world problems. Emphasis on Python, data visualization and exploratory analysis, gaining hands-on experience with tools and techniques that support the entire data pipeline—from data wrangling to communication of insights—preparing me to operate at the intersection of analytics, technology and strategy.
MSc in Genetics and Molecular Biology [Emphasis in Bioinformatics] (2024)
Federal University of Pernambuco (UFPE), Recife, Pernambuco, Brazil
Developed AMP-Identifier, a machine learning-based tool for genome mining focused on the discovery of bioactive molecules. Conducted the first comprehensive characterization of defensins in R. communis, integrating genomic, transcriptomic and structural biology analyses.
BSc in Biomedical Sciences [Emphasis in Clinical Pathology and Bioinformatics] (2022)
Federal University of Pernambuco (UFPE), Recife, Pernambuco, Brazil
Gained practical experience in hospital laboratories and molecular biology research environments. Further specialized in computational biology through a bioinformatics internship at academic research centers. Undergraduate thesis focused on the eIF4E gene family and the impact of mutations on protein synthesis and susceptibility to viruses that hijack the host translation machinery.
Certifications
Publications
[4] M. V. F. Ferraz et al., Association strength of E6 to E6AP/p53 complex correlates with HPV‐mediated oncogenesis risk, Biopolymers, vol. 113, no. 10, p. e23524, 2022. DOI: 10.1002/bip.23524
[3] L. M. B. Vilela et al., Approaches for Identification and Validation of Antimicrobial Compounds of Plant Origin: A Long Way from the Field to the Market, in Eco-Friendly Biobased Products Used in Microbial Diseases, CRC Press, 2022, pp. 183–222. DOI: 10.1201/9781003243700
[2] R. C. C. da Silva et al., Omics-driven bioinformatics for plant lectins discovery and functionalannotation – A comprehensive review, International Journal of Biological Macromolecules, p. 135511, 2024. DOI: 10.1016/J.IJBIOMAC.2024.135511
[1] F. A. de Andrade and M. A. de Luna-Aragão et al., Deciphering Cowpea Resistance to Potyvirus: Assessment of eIF4E Gene Mutations and Their Impact on the eIF4E-VPg Protein Interaction, Viruses, 2025. DOI: 10.3390/v17081050
Gallery
A visual collection of my work, projects, and scientific renderings. From molecular visualizations to computational outputs, this gallery showcases the intersection of art and science in bioinformatics research.
ESP from eIFEs and ESP
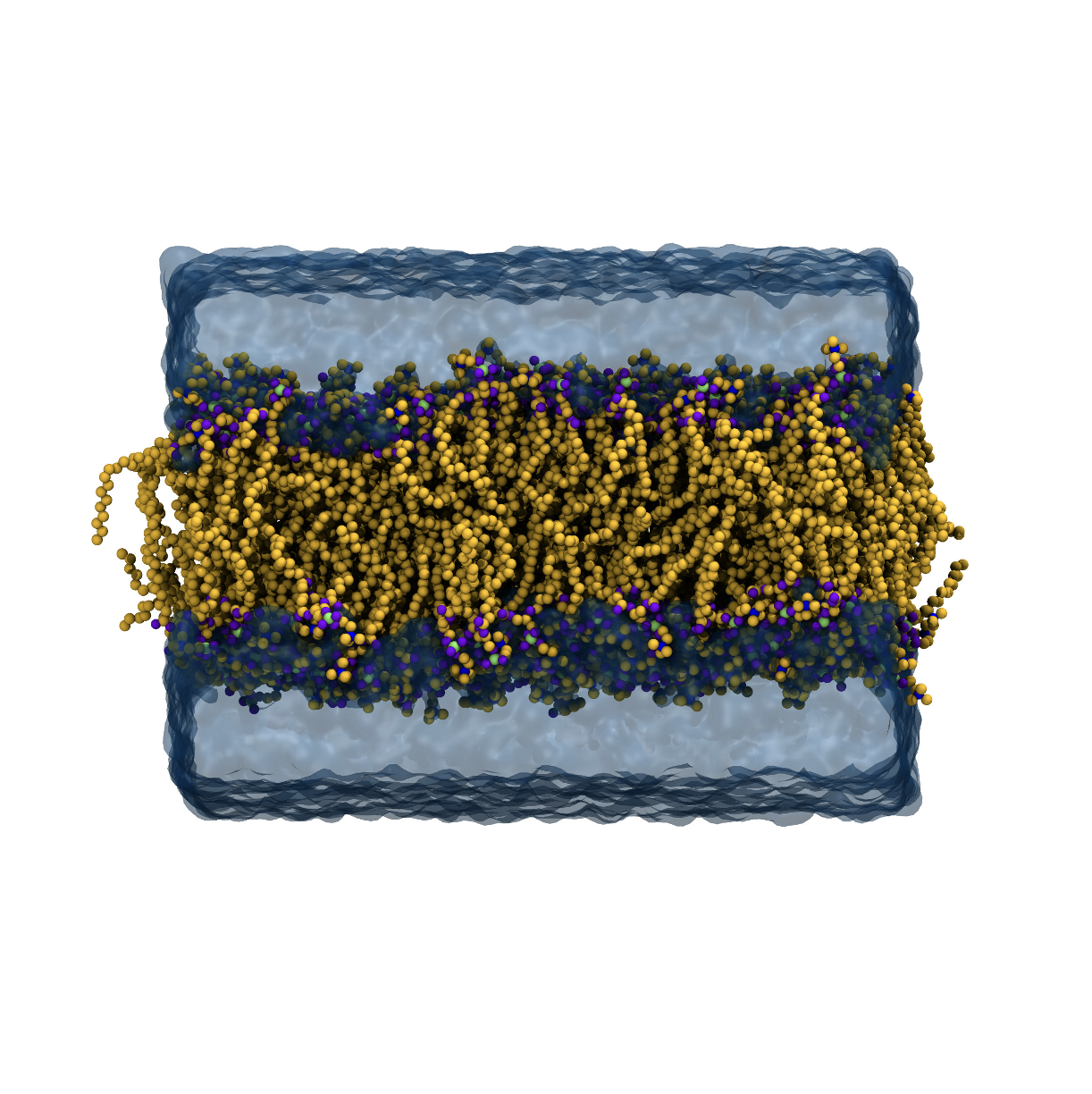
DPOC membrane
Data Visualization
Genomic Analysis
Machine Learning Model
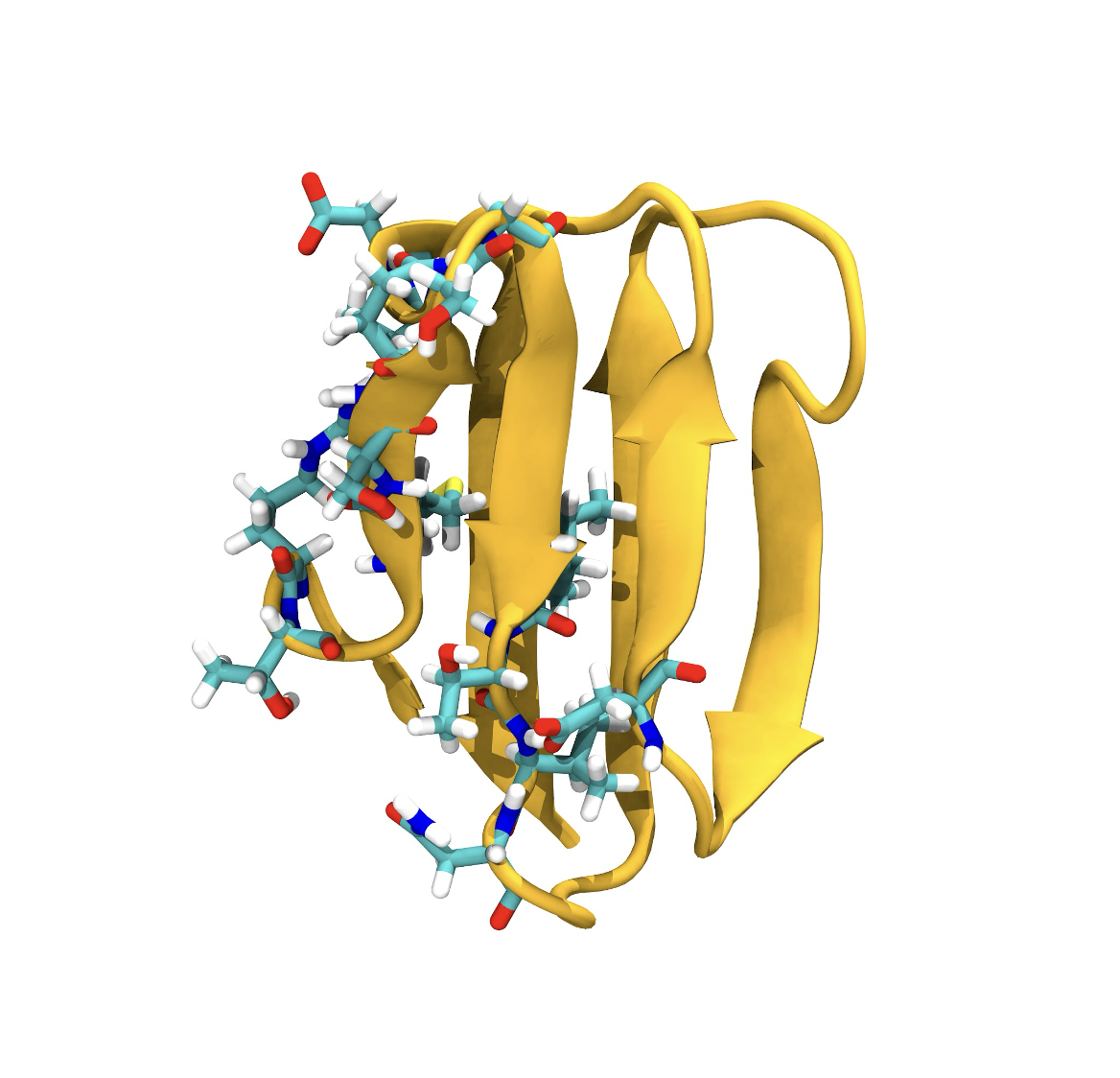
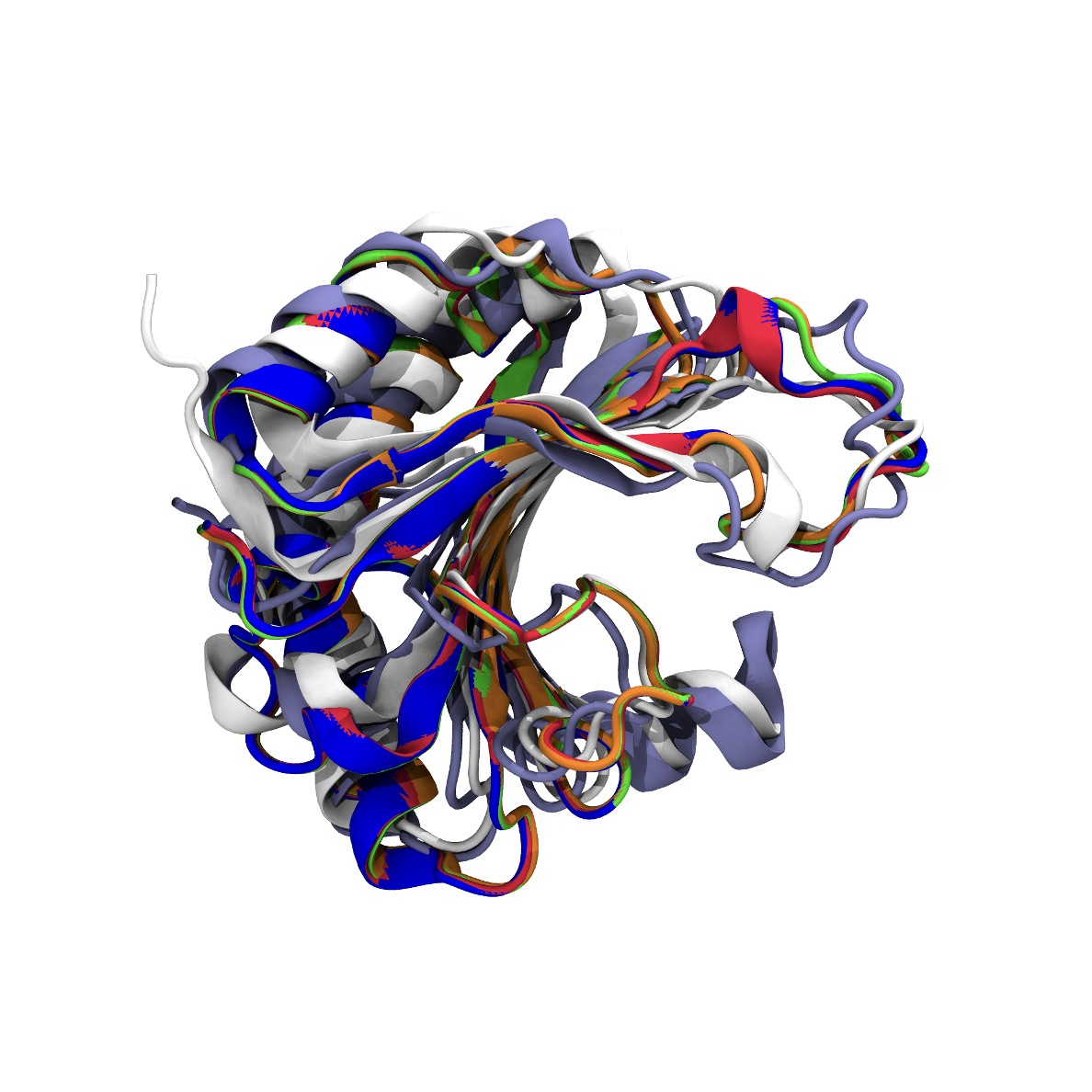
Structural Biology
Conference Presentations & Invited Talks
Teaching Experience
Awards, Recognitions & Achievements
Code/Scripts
A curated vault of open-source code, pipelines and experiments I use in my analyses and projects is available for public access. Enter below to explore the repository:
C:\REPOSITORY\code_
Materials
If you have arrived here after attending one of my courses, classes, or lectures, please feel free to access the cards below. These cards contain all the materials used during the sessions, including presentations, examples, and recommended articles. Thank you for your interest, and I hope this material aids you in your knowledge journey. Feel free to contact me, you can find the contact information in the Contact Section.
Lecture notes are stored in the digital archives. Click below to enter the directories.
C:\DOCUMENTS\sharing_
BioHub
The BioHub / Signal Hub is also available as a Python CLI (Command Line Interface) version, providing a comprehensive hub of tools for structural bioinformatics analyses. The website offers additional features including automated graph generation and 3D protein visualization. Both tools are currently under active development. Since the project is in its beta phase, feedback and contributions are highly encouraged to enhance its capabilities. Feel free to contact me for any suggestions or improvements! You can found the contact information in the Contact Section.
A collection of bioinformatics tools for structural biology analyses. Click below to enter and explore the tools (Still in Beta version, feedbacks are welcome!):
BioHub / Signal Hub Website_
B101NF0
Featured Videos (This is a personal curation of media I've watched and loved... If you geek out on structural bio and data science like I do, I hope you should enjoy this!)
The Thinking Game takes you on a journey into the heart of DeepMind, capturing a team striving to unravel the mysteries of intelligence and life itself. Filmed over five years by the award winning team behind AlphaGo, the documentary examines how Demis Hassabis’s extraordinary beginnings shaped his lifelong pursuit of artificial general intelligence. It chronicles the rigorous process of scientific discovery, documenting how the team moved from mastering complex strategy games to the ups and downs of solving a 50-year-old "protein folding problem" with AlphaFold.
Personal Disclaimer:
AlphaFold (AF2/AF3) represents one of the most remarkable scientific advances of recent years, and witnessing its development has been like watching a flower bloom for the first time, especially for anyone working in structural bioinformatics. That said, it is essential to remember that the protein folding problem remains far beyond what we currently understand in thermodynamics, classical mechanics, and what deep learning models can truly access. AF3 did not “solve” protein folding, although it undeniably marks a significant step forward.
It is important to acknowledge the broader scientific context (But here, I have also a another (very) personal disclaimer):
AF2 enabled an extraordinary leap forward largely because it was openly released, empowering the global community to build upon it. Unfortunately, AF3 shifted toward a closed model, limiting access and slowing the collective progress of the field. Only open science can ensure a fair, transparent, and inclusive future for research.
In this first part of the Hello Nextflow training course, you ease into the topic with a very basic domain-agnostic Hello World example, which you'll progressively build up to demonstrate the usage of foundational Nextflow logic and components. To know more about the Nextflow training, visit the official Hello Nextflow website.
This is the story of the world's most beloved programming language: Python. What began as a side project in Amsterdam during the 1990s became the software powering artificial intelligence, data science and some of the world’s biggest companies. But Python's future wasn't certain, at one point it almost disappeared. This 90-minute documentary features Guido van Rossum, Travis Oliphant, Barry Warsaw, and many more, and they tell the story of Python’s rise, its community-driven evolution, the conflicts that almost tore it apart, and the language’s impact on... Well… Everything!!! To know more about Python, visit the official website.
The rapidly evolving field of protein design is revealing solutions to some of the world’s greatest problems, whether it's blocking a virus, breaking down a pollutant or creating brand-new materials. In conversation with TED’s Whitney Pennington Rodgers, biochemist David Baker explores his team’s Nobel Prize-winning work using AI to design new proteins with functions never before seen in nature — achieving breakthroughs that have fundamentally changed the future of science. This conversation was part of an exclusive TED Membership event. TED Membership is the best way to support and engage with the big ideas you love from TED. To learn more, visit the TED official website.
This video covers Bindcraft, a one-shot design of functional binders. Starting with a brief overview of current binder design tools and conceptually explaining the Bindcraft's central hallucination process that engages loss and confidence metrics to predict sequence from structure. Cho then delves into the internal workflow of Bindcraft describing AF2 Mulitmer for backbone and sequence codesign, sequence optimization with MPNNsol M, and validation/filtering with AF2Monomer. The successes and limitations of this tool are addressed.
The biggest problems in the world might be solved by tiny molecules unlocked using AI.
This is the inside story of how David Baker, Demis Hassabis and John Jumper won the 2024 Nobel Prize in Chemistry for advances in computer-assisted protein design and structure prediction. Proteins are biological nano-machines that perform a vast array of vital functions inside of every living.
Satya Nadella on: Why he doesn’t believe in AGI but does believe in 10% economic growth; Microsoft’s new topological qubit breakthrough and gaming world models; Whether Office commoditizes LLMs or the other way around… And much more!
AI models are trained and not directly programmed, so we don’t understand how they do most of the things they do. With two new papers, Anthropic's researchers have taken significant steps towards understanding the circuits that underlie an AI model’s thoughts. In one example from the paper, they find evidence that Claude will plan what it will say many words ahead, and write to get to that destination. They show this in the realm of poetry, where it thinks of possible rhyming words in advance and writes each line to get there. This is powerful evidence that, even though models are trained to output one word at a time, they may think on much longer horizons to do so. Read more: Tracing the thoughts of a large language model
ESM3 is a generative language model for programming biology. In experiments, researchers found ESM3 can simulate 500M years of evolution to generate new fluorescent proteins. Read more: Simulating 500 million years of evolution with a language model
When Thomas Wagner’s son received a diagnosis of Alexander disease, a rare genetic disorder, he set out to learn everything he could. Through these conversations, Thomas has helped jumpstart new research into potential treatmentsd. He realized there might be similarities between Alexander disease, which had very little active research, and other diseases with much more funding and ongoing research. He began to reach out to those researchers, scientists and doctors, many of whom had never heard of Alexander disease before. Through these conversations, Thomas has helped jumpstart new research into potential treatments. Find out more about Thomas' story on Googles' blog: Finding answers, building hope
